How to Detect COVID-19 Variants of Concern
How to Detect COVID-19 Variants of Concern It’s a little deja-vu writing this title one year after a similar blog post on how to validate a COVID-19 assay at the start of the pandemic. In many ways, the challenges are similar: limited reagents/control material, and rising case counts. At least now, there is increasing support in the way of funding from the federal government that could help with monitoring and surveillance. I’m going to summarize the current methods available for detecting the Variants of Concern and emerging variants. Whole Genome Sequencing The principle method used by many is whole genome sequencing. It has the advantage of being able to comprehensively examine every letter (nucleotide) of the SARS-CoV-2 genome (30 kilobases long). At our institution, I’ve been working on the effort to sequence all of our positive specimens. While it is achievable, it is not simple nor feasible at most locations. Limitations include:
Financial: must already own expensive sequencers
Expertise: advanced molecular diagnostics personnel needed who perform NGS testing
Data Analytics: Dbioinformatics personnel needed to create pipelines, analyze data and report it in a digestible format.
Timing: the process usually takes a week at best and several weeks if there is a backlog or not enough samples for a sequencing run to be financially viable.
Sensitivity: the limit of detection for NGS is 30 CT cycles, which for us includes only about 1/2- 1/3 of all positive COVID19 specimens.
Bottom line: WGS is the best at detecting new/ emerging strains or mutations when cost/ time is not a concern.
Mutation Screening
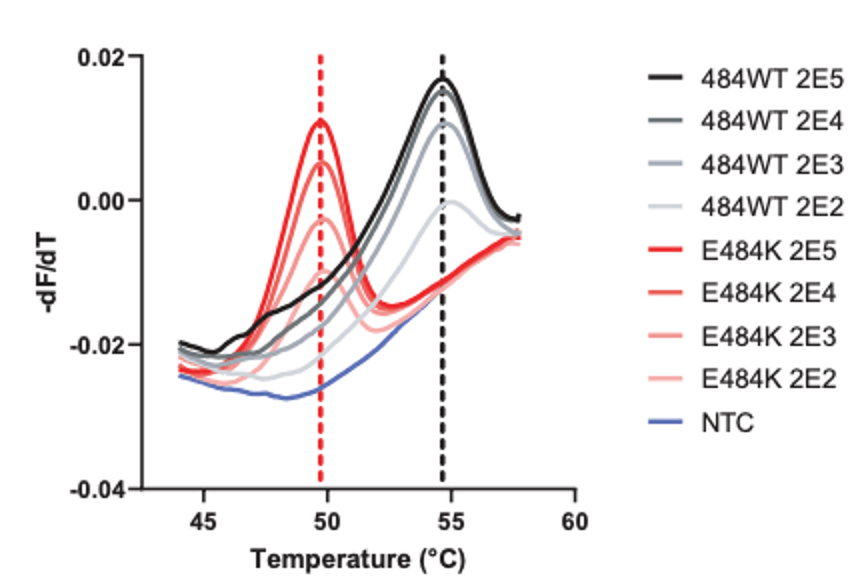
Other institutions have begun efforts to screen for variants of concern by detecting characteristic mutations. For instance, the N501Y mutation in the spike protein is common to the major Variants of Concern (UK B.1.1.7, Brazil P.1, and S Africa B.1.351) and E484K is present in the Brazil (P.1), S Africa (B.1.351) and New York Variant (B.1.526). Thus, several institutions (listed below) took approaches to 1) screen for these mutations and then 2) perform WGS sequentially.
Subscribe to our
Newsletter
***We Promise, no spam!